Overview
The Koelle Laboratory studies immune responses to infections, pathogen genetic variation, and the relationship between host genomics and infection severity.
Pathogens of interest include herpes simplex viruses types 1 and 2, varicella zoster virus, vaccinia (the vaccine agent for smallpox), Merkel cell polyoma virus (MCPyV), and Mycobacterium tuberculosis (MTB).
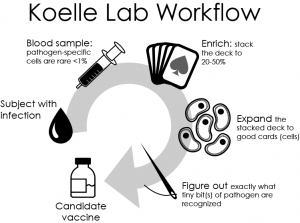
These agents mostly have large genomes, so determination of the antigens that drive T-cell responses is a non-trivial problem. Their specific technical expertise is in the use of genomic libraries and genome-spanning ORF sets to interrogate CD8 and CD4 T-cell responses to a very high level of definition.
A suite of modern immunology tools such as intracellular cytokine cytometry, tetramers, cell killing assays, etc. are used in this work. Candidate HSV-1 and HSV-2 vaccine candidates have been identified, and some have been studied in mice and are poised to enter phase I trials. The laboratory is decoding the T cell response to MCPyV to assist cancer therapy.
A specific focus of the lab has been measuring cellular immunity at sites of infection, such as skin, the female genital tract, the cornea, trigeminal ganglia, and tumor biopsies. Homing markers identified with this approach are being studied at the mechanistic level to understand how virus-specific T-cells are programmed to rapidly return to these sites.
Newer initiatives include studies of T-cell diversity using regular and deep sequencing of TCR hypervariable regions, studies of HSV-1 and HSV-2 diversity at the DNA level, and detailed study of long-term T-cell memory decades after pathogen exposure. Large numbers of T-cell tests for MTB exposure are being run in the clinical lab and epitope discovery has begun for MTB.
Fellows interested in modern laboratory immunology are welcome.
Principal investigator
Diseases we study
- Herpes Simplex Virus
- Malaria
- Merkel Cell Carcinoma & Merkel Cell Polyoma Virus
- Newcastle Disease
- Smallpox & Other Orthopoxviruses
- T Cell Epitope Discovery
- T Cell Receptor Biology
- Tuberculosis
- Varicella Zoster Virus
- Zika Virus